|
|
| (289 intermediate revisions by 2 users not shown) |
| Line 1: |
Line 1: |
| − | <div id='mainbackground'></div> | + | <div class="row"> |
| − | <div id="mw-content-text" lang="en" dir="ltr" class="mw-content-ltr"><div class="mw-parser-output"><div class="jumbotron"> | + | <div class="col-md-offset-0 col-md-15"> |
| − | <h1><span class="mw-headline" id="Tweeki"></span>What We Do</h1> | + | <div class="jcarousel-wrapper"> |
| − | <p>Our lab works on problems in the fields of computational biology and bioinformatics. We have studied a wide variety of topics in cancer genomics, gene regulation, and molecular evolution, with particular focuses in post-transcriptional regulation and tumor evolution.</p>
| + | <div class="jcarousel jcarousel-randomize"> |
| | + | <div class="jcarousel-list"> |
| | + | <div class="jcarousel-item"> |
| | + | {{banner|direction=right|title=Welcome |section= |section-link=Chuang_Lab|image=clab_banner2.jpg|width=20%|quote=Our lab uses computational and mathematical approaches to investigate the mechanisms that govern cancer cells and their spatial ecosystems. The lab specializes in problems in cancer image analysis, genomics, and evolution.}} |
| | + | </div> |
| | + | <div class="jcarousel-item"> |
| | + | {{banner|direction=right|title=Research Topics|section=|image=PDXpicture.png |width=15%|quote=The lab uses computational and mathematical approaches to understand cancer cells and their spatial ecosystems. We develop and apply techniques from a variety of disciplines, including deep neural networks, genomic and evolutionary analysis, and biophysical modeling. We are currently focused in: 1) Multiplex and histopathological cancer image analysis using deep learning, and 2) Cancer evolution in response to therapy.}} |
| | + | </div> |
| | + | <div class="jcarousel-item"> |
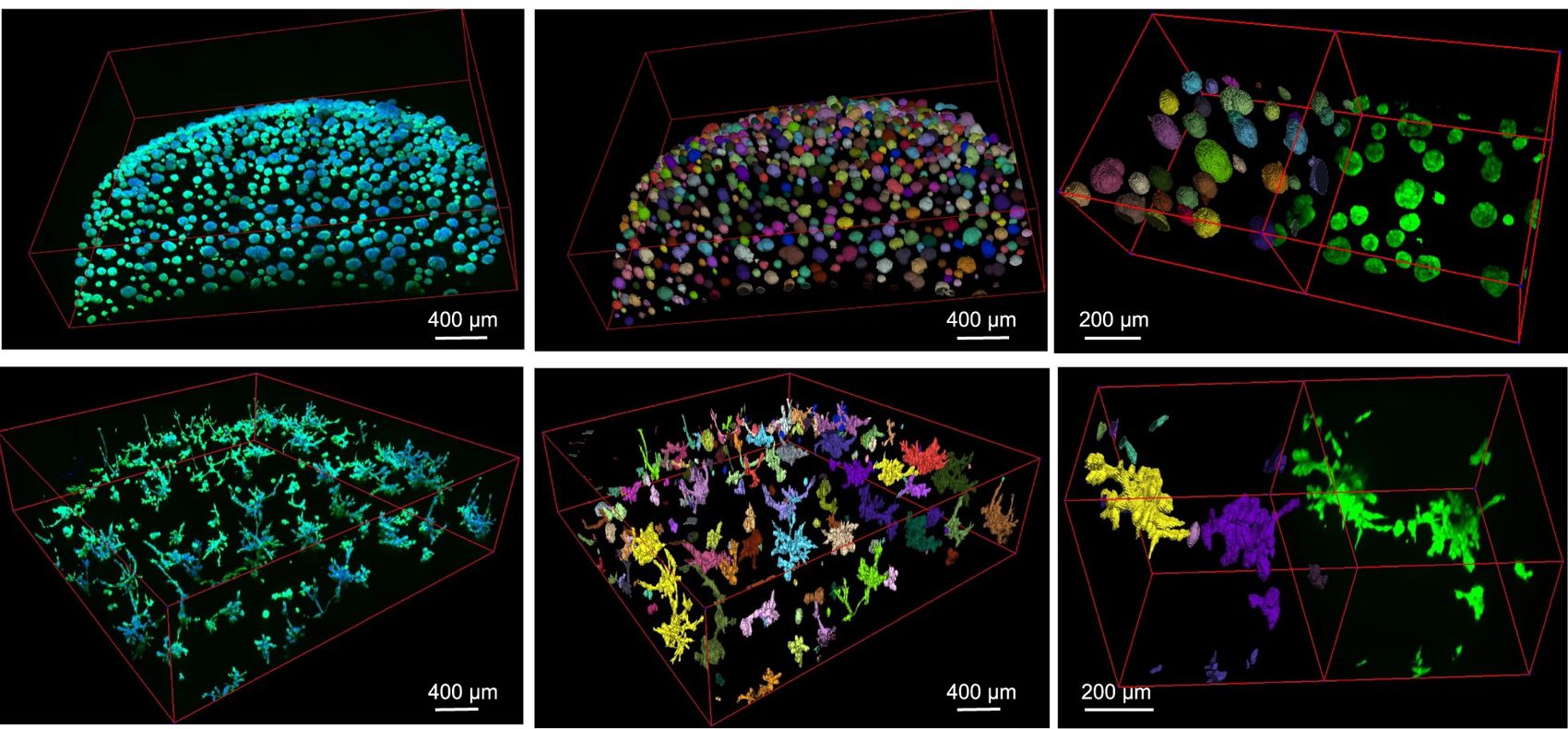
| | + | {{banner|direction=left|title = Publications | section= [https://link.springer.com/article/10.1038/s41467-023-44162-6 Cellos: deconvolution of 3D organoid dynamics at cellular resolution]|image= CellosFig3.jpg|width=45%|quote=}} |
| | + | </div> |
| | + | <div class="jcarousel-item"> |
| | + | {{banner|direction=right|title=People|section= |image=East_rock_lookout_2024.jpg|width=15%|quote=Meet the members of the Chuang lab}} |
| | + | </div> |
| | + | <div class="jcarousel-item"> |
| | + | {{banner |
| | + | |direction=right |
| | + | |title = News |
| | + | |section = [http://chuanglab.org/News PDXNet U24 grant] |
| | + | |image=jeff-chuang-9922mirrored.jpg |
| | + | |width=45% |
| | + | |quote= The lab has been awarded a National Cancer Institute grant to continue our coordination of data sharing and analysis to general cancer clinical trials for the PDXNet. We are working with institutes including MD Anderson, Dana Farber, Huntsman Cancer Institute, Baylor College of Medicine, the Wistar Institute, Washington University, UC Davis, and Virginia Commonwealth University. For this project we will be analyzing hundreds of new and existing PDX samples toward the goal of developing a clinical trial based on patient-derived xenografts.}} |
| | + | </div> |
| | + | </div> |
| | </div> | | </div> |
| | + | <span class="jcarousel-control-prev">[[#|‹]]</span> |
| | + | <span class="jcarousel-control-next">[[#|›]]</span> |
| | + | <p class="jcarousel-pagination"></p> |
| | + | </div> </div> |
| | + | |
| | <div class="row-fluid clearfix" style="width:90%;margin:auto;text-align:center"> | | <div class="row-fluid clearfix" style="width:90%;margin:auto;text-align:center"> |
| | <div class="col-md-4"> | | <div class="col-md-4"> |
| − | <h2><span class="mw-headline" id="Contact Information"><span class="fa fa-news"></span></span> Contact Information</h2> | + | <h2><span class="mw-headline" id="Research"><span class="fa fa-news"></span></span> Research</h2> |
| − | <p>Prof. Jeffrey Chuang <br> | + | <p>Topics that the Chuang Lab investigates |
| − | The Jackson Laboratory for Genomic Medicine <br>
| |
| − | 10 Discovery Drive <br>
| |
| − | Farmington, CT 06032 <br>
| |
| − | jeff (dot) chuang (at) jax (dot) org
| |
| − | <br>
| |
| − | <br>
| |
| − | Administrative assistant: Julie Yarmolovich <br>
| |
| − | julie (dot) yarmolovich (at) jax (dot) org<br>
| |
| | </p> | | </p> |
| − | <btn>Jeffrey Chuang</btn> | + | <btn>Research Topics</btn> |
| | </div> | | </div> |
| | <div class="col-md-4"> | | <div class="col-md-4"> |
| Line 26: |
Line 46: |
| | </div> | | </div> |
| | <div class="col-md-4"> | | <div class="col-md-4"> |
| − | <h2><span class="mw-headline" id="Features.3F"><span class="fa fa"></span></span> Features?</h2> | + | <h2><span class="mw-headline" id="Openings"><span class="fa fa"></span></span> Openings</h2> |
| − | <p>Most of the things that are possible with <a rel="nofollow" class="external text" href="http://getbootstrap.com">Bootstrap</a> are possible with Tweeki. | + | <p>Student/postdoctoral/research scientist openings in cancer computational biology. |
| | </p> | | </p> |
| − | <div class="btn-group"><a href="/wiki/Components" id="n-Components" class="btn btn-default">See how to do it »</a></div> | + | <btn>Openings</btn> |
| | </div> | | </div> |
| | </div> | | </div> |
| Line 35: |
Line 55: |
| | </p><p><br /> | | </p><p><br /> |
| | </p> | | </p> |
| − |
| |
| − | [[File:jax-logo.jpg|class=img-responsive]]
| |
| − |
| |
| − | [[Research Topics]] <div>
| |
| − | <div>[[Publications]]
| |
| − | <div>[[People]]
| |
| − | <div>[[Jeffrey Chuang]]
| |
| − | <div> [[News]]
| |
| − | <div>[[Links]]
| |
| − |
| |
| − |
| |
| − | <div id='mainbox1' class='leftborder gy' style='float:right'>
| |
| − | <h2>Contact Info</h2>
| |
| − | Prof. Jeffrey Chuang <br>
| |
| − | The Jackson Laboratory for Genomic Medicine <br>
| |
| − | 10 Discovery Drive <br>
| |
| − | Farmington, CT 06032 <br>
| |
| − | jeff (dot) chuang (at) jax (dot) org
| |
| − | <br>
| |
| − | <br>
| |
| − | Administrative assistant: Julie Yarmolovich <br>
| |
| − | julie (dot) yarmolovich (at) jax (dot) org<br>
| |
| − | </div>
| |
| − | <br>
| |
| − | <div id='mainbox2' class='leftborder oy' style='float:left;margin-top:19em'>
| |
| − |
| |
| − | __NOTOC__
| |
| − | <div id='mainbox1' class='leftborder gy' style='float:right'>
| |
| − | <h2>Contact Info</h2>
| |
| − | Prof. Jeffrey Chuang <br>
| |
| − | The Jackson Laboratory for Genomic Medicine <br>
| |
| − | 10 Discovery Drive <br>
| |
| − | Farmington, CT 06032 <br>
| |
| − | jeff (dot) chuang (at) jax (dot) org
| |
| − | <br>
| |
| − | <br>
| |
| − | Administrative assistant: Julie Yarmolovich <br>
| |
| − | julie (dot) yarmolovich (at) jax (dot) org<br>
| |
| − | </div>
| |
| − | <br>
| |
| − | <div id='mainbox2' class='leftborder oy' style='float:left;margin-top:19em'>
| |
| − |
| |
| − | <h2>Introduction</h2>
| |
| − | Our lab works on problems in the fields of computational biology and bioinformatics. We have studied a wide variety of topics in cancer genomics, gene regulation, and molecular evolution, with particular focuses in post-transcriptional regulation and tumor evolution.
| |
| − | </div>
| |
| − |
| |
| − | <!-- <div id='mainbox3' class='leftborder yb' style='float:left; margin-top:1em'> -->
| |
| − | <!-- <h2>Research Openings</h2> -->
| |
| − | <!-- </div> -->
| |
| − |
| |
| − | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br>
| |
| − | <br><br><br><br><br><br>
| |
| − | [[Research Topics]] <div>
| |
| − | [[Publications]]
| |
| − | [[People]]
| |
| − | [[Links]]
| |
| − |
| |
| − | ==Chuang Lab Awarded U24 for Data Coordination Center for NCI PDXNet (October 2017)==
| |
| − | Together with Seven Bridges Genomics, we have been awarded a National Cancer Institute grant to coordinate data sharing and analysis for the PDXNet. We are working with esteemed collaborators at institutes including MD Anderson, the Huntsman Institute at the University of Utah, Baylor College of Medicine, the Wistar Institute, and Washington University. For this project we will be analyzing hundreds of new and existing PDX samples toward the goal of developing a clinical trial based on patient-derived xenografts.
| |
| − | https://www.jax.org/news-and-insights/2017/october/jax-seven-bridges-cancer-data-platform
| |
| − | <br>
| |
| − |
| |
| − | ==Work on cancer evolution is published in Nature Genetics==
| |
| − | Our project led by Dr. Javad Noorbakhsh quantifying the ability of TCGA data to reveal evolutionary dynamics has been published.
| |
| − | https://www.nature.com/ng/journal/v49/n9/full/ng.3876.html
| |
| − | <br>
| |
| − |
| |
| − | ==Postdoctoral / Research Scientist Opening in Cancer Computational Biology (June 2016)==
| |
| − |
| |
| − | The Chuang Lab at JAX-GM is seeking scientists to join our projects in cancer genomics and evolution. We are a computational lab that works closely with multiple experimental colleagues to study tumor evolution and resistance to therapy. We commonly compare treated and untreated samples through analysis of sequencing data (exome, RNA-seq, single cell, targeted sequencing, etc.). Our projects take advantage of JAX’s strengths in patient-derived xenografts, a powerful model system in which human tumors are engrafted and studied in immunocompromised mice. JAX has developed >400 such tumors, which we are characterizing extensively via genomics and clinical perturbations at an institutional level. New team members will have unique opportunities to investigate mechanisms of cancer from these types of data, to design new studies, and to take advantage of cloud computing approaches for large scale data analysis. Particular areas of emphasis include gastric cancer, breast cancer, and immunotherapy. The ideal candidate will have prior expertise in cancer genomics, computational biology, or molecular evolution. Outstanding candidates transitioning from another quantitative discipline such as applied math, physics, or computer science will be considered, though they would be expected to be highly motivated and rapidly become expert in cancer genomics. Strong computing (programming, large scale data analysis, statistical inference, etc), publication, and communication skills are essential. Interested applicants should send a CV and a research statement to Jeffrey Chuang at jeff.chuang@jax.org. Reference letters will be requested subsequently. See also the general comments on openings below.
| |
| − |
| |
| − | ==Chuang Lab Profile (June 2016)==
| |
| − | We have had the honor of being profiled for episode 1 of JAX's Supplemental Material, an informal podcast series explaining research at JAX.
| |
| − | https://soundcloud.com/jax-supplemental-material
| |
| − | <br>
| |
| − |
| |
| − | Also, the joint work of our lab with the Graveley lab at U. Conn has been featured in [https://www.jax.org/news-and-insights/2016/april/jax-uconn-health-teaming-up-to-improve-human-health?utm_source=LeadingTheSearch%20&utm_medium=Email&utm_campaign=LeadingTheSearch%204.21&mkt_tok=eyJpIjoiT1Rrd016QTJNRGRoWXpNMyIsInQiOiI5OFZobFA5akZrRGY5OURVazBSdHVjdDFVMFlQdzg4TWpwUHhMaFVWeUJseEdQVlNUbVJRQVBpb3hhVEFCWk5QN0FRS0x2enltNUJkMVFnVzYycEJXQStQa25WWGphNldHNmgyM2k3WFwvZkk9In0%3D The Search].
| |
| − |
| |
| − | Our work in Big Data Genomics has also been recently profiled [https://www.jax.org/news-and-insights/2016/may/jeff-chuang-is-on-the-front-lines-of-big-data here]
| |
| − | <br>
| |
| − |
| |
| − | ==Chuang Lab Awarded NCI Supplement for Cloud Computing (February 2016)==
| |
| − | Our lab, in collaboration with Seven Bridges Genomics, has been awarded a National Cancer Institute supplementary grant to investigate tumor heterogeneity using TCGA data on the Amazon cloud. We presented this work in the NCI Cloud Pilots session at AACR in April 2016.
| |
| − | <br>
| |
| − |
| |
| − | ==Big Genomic Data Skills Training for Professors (September 2015)==
| |
| − | Prof. Reinhard Laubenbacher (JAX/UCHC), Dr. Charlie Wray (JAX), and Prof. Chuang are leading a course in summer 2016 to provide training in big data genomics to professors from smaller colleges and regional universities. A description of the course is here. The next iteration will be given in Bar Harbor, ME in summer 2017.
| |
| − | https://www.jax.org/news-and-insights/2016/january/jax-offers-big-genomic-training-for-professors-course
| |
| − | <br>
| |
| − |
| |
| − | ==Chuang Lab Awarded NIH R21 (July 1, 2015)==
| |
| − | Our lab, in collaboration with Profs. Kyuson Yun and Craig Nelson, has been awarded a new R21 grant from the Dissection of Tumor Evolution Using Patient-Derived Xenografts!
| |
| − | <br>
| |
| − |
| |
| − | ==Work on cancer structural mutations is published in BMC Genomics==
| |
| − | Our project led by Dr. Krzysztof Grzeda, and in collaboration with the labs of Prof. Ed Liu and Prof. Axel Hillmer has been published.
| |
| − | http://www.biomedcentral.com/1471-2164/15/1013/abstract
| |
| − |
| |
| − | ==Work on ribosomal stalling and neurodegeneration is published in Science (July 25, 2014)==
| |
| − | Our collaboration with Prof. Sue Ackerman, with the informatics work led by Ivan Dotu, has been published in Science.
| |
| − | http://www.sciencemag.org/content/345/6195/455.full
| |
| − |
| |
| − | ==Chuang Lab Awarded NIH R21 (January 27, 2014)==
| |
| − | Our lab, in collaboration with Prof. Brenton Graveley and Research Assistant Professor Dr. Ivan Dotu, has been awarded a new R21 grant from the NIH!
| |
| − | <br>
| |
| − | http://www.eurekalert.org/pub_releases/2014-01/jl-jgm012714.php
| |
| − |
| |
| − | ==Notes for Graduate Students (Updated July 2014)==
| |
| − | The Chuang Lab is accepting graduate students through the UCHC program in Developmental Biology and Genetics. We are looking to hire 1-2 new graduate students for students applying in 2014, to join the program in Fall 2015. If you have interest in our lab, please contact Prof. Chuang directly. General information on graduate studies with JAX is available at: <br>
| |
| − | http://education.jax.org/predoc/
| |
| − | <br>
| |
| − | Students working with the Chuang Lab should apply through the UCHC admissions process. Information on that is available at: <br>
| |
| − | http://grad.uchc.edu/prospective/programs/phd_biosci/concentration/devbio_genetics/index.html
| |
| − |
| |
| − | ==Openings==
| |
| − |
| |
| − | Our lab is always open to new students and postdocs excited to work on computational biology problems here at the Jackson Laboratory for Genomic Medicine. We are a computational biology lab specializing in cancer genomics, post-transcriptional regulation and molecular evolution. Our lab works closely with a number of experimental labs to analyze and interpret high-throughput functional genomic data from human and mouse model systems.
| |
| − |
| |
| − | At the post-doctoral level, well-suited candidates should have Ph.D. experience in computational biology, applied math, physics, computer science, or a similar quantitative discipline. Outstanding applicants new to computational biology will also be considered. Strong computing (programming, large scale data analysis, statistical inference, etc), publication, and communication skills are essential. At the graduate student level, a variety of backgrounds can fit, but enthusiasm and aptitude for computational biology are what define the lab.
| |
| − |
| |
| − | Hirees will be joining the Jackson Laboratory for Genomic Medicine, a research institute dedicated to improving human health through genomic approaches. We moved into our newly constructed building and lab space in October 2014. The institute is in Farmington, CT adjacent to the University of Connecticut Medical School and in the Hartford metropolitan area. This institute is part of a $1.1 billion new venture between the Jackson Laboratory (based in Bar Harbor, ME), which is one of the world's leaders in mouse genetics, and the state of Connecticut. JAX Genomic Medicine is building an institute of 30 scientific research groups in the next few years, one third of which are expected to be in computational biology. New hirees will have the opportunity to join one of the first computational biology labs in the institute. Hartford is a lively metropolitan area of over one million people and is within a two hour drive of both Boston and New York City.
| |
| − |
| |
| − |
| |
| − | Interested applicants should send a CV and a research statement to Jeffrey Chuang at jeff.chuang@jax.org. Reference letters will be requested subsequently.
| |
| − |
| |
| − | Jackson Lab website: http://www.jax.org/
| |
| − | Bioscience in Connecticut website: http://biosciencect.uchc.edu/jackson_laboratory/index.html
| |
| − |
| |
| − | <br><br>
| |
| − | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/>
| |