Difference between revisions of "Chuang Lab"
| (186 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <div class="row"> | ||
| + | <div class="col-md-offset-0 col-md-15"> | ||
<div class="jcarousel-wrapper"> | <div class="jcarousel-wrapper"> | ||
<div class="jcarousel jcarousel-randomize"> | <div class="jcarousel jcarousel-randomize"> | ||
<div class="jcarousel-list"> | <div class="jcarousel-list"> | ||
<div class="jcarousel-item"> | <div class="jcarousel-item"> | ||
| − | {{banner|direction=right|title=|section= | + | {{banner|direction=right|title=Welcome |section= |section-link=Chuang_Lab|image=clab_banner2.jpg|width=20%|quote=Our lab uses computational and mathematical approaches to investigate the mechanisms that govern cancer cells and their spatial ecosystems. The lab specializes in problems in cancer image analysis, genomics, and evolution.}} |
</div> | </div> | ||
| − | <div class="jcarousel-item"> | + | <div class="jcarousel-item"> |
| − | {{banner|direction=left|title= | + | {{banner|direction=right|title=Research Topics|section=|image=PDXpicture.png |width=15%|quote=The lab uses computational and mathematical approaches to understand cancer cells and their spatial ecosystems. We develop and apply techniques from a variety of disciplines, including deep neural networks, genomic and evolutionary analysis, and biophysical modeling. We are currently focused in: 1) Multiplex and histopathological cancer image analysis using deep learning, and 2) Cancer evolution in response to therapy.}} |
| + | </div> | ||
| + | <div class="jcarousel-item"> | ||
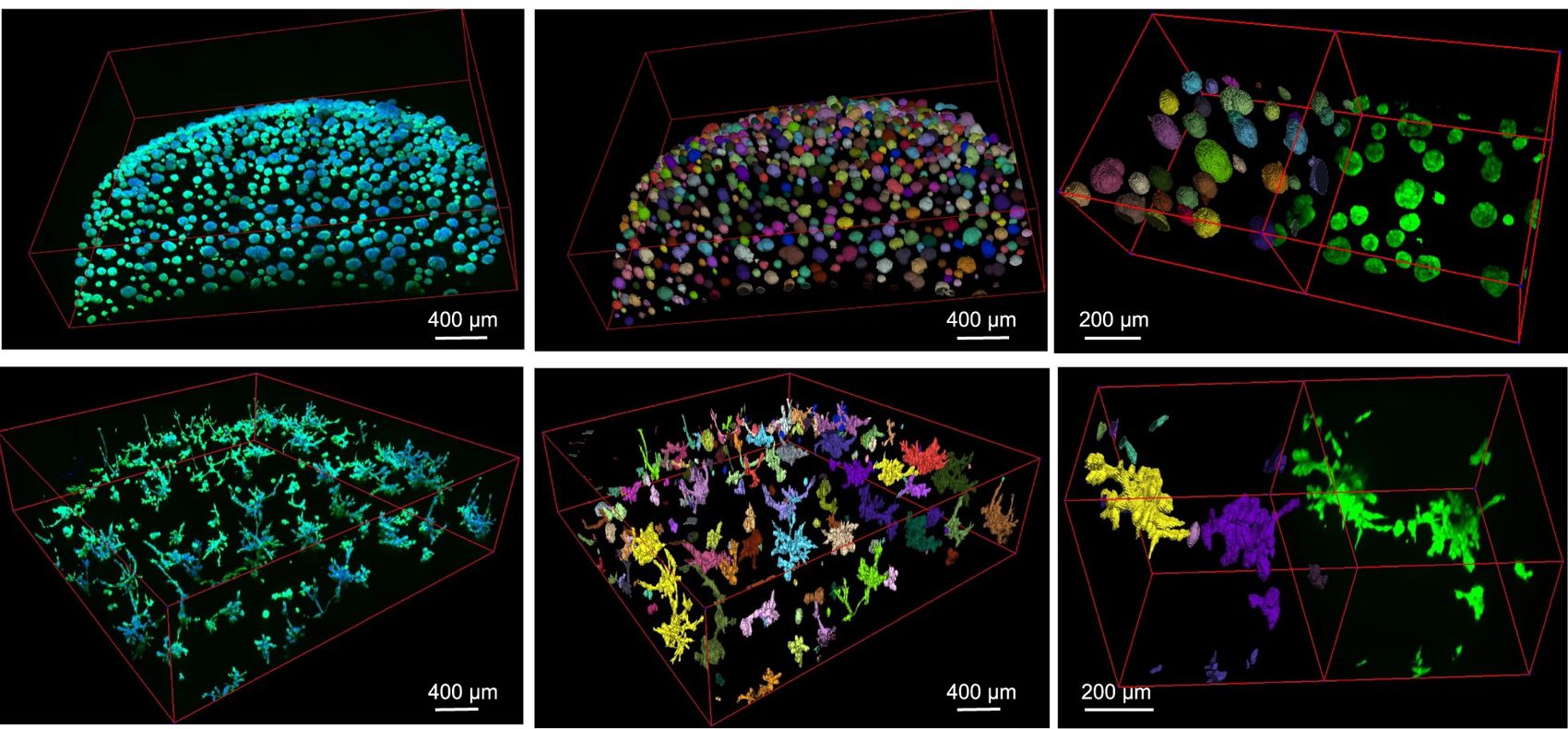
| + | {{banner|direction=left|title = Publications | section= [https://link.springer.com/article/10.1038/s41467-023-44162-6 Cellos: deconvolution of 3D organoid dynamics at cellular resolution]|image= CellosFig3.jpg|width=45%|quote=}} | ||
| + | </div> | ||
| + | <div class="jcarousel-item"> | ||
| + | {{banner|direction=right|title=People|section= |image=East_rock_lookout_2024.jpg|width=15%|quote=Meet the members of the Chuang lab}} | ||
</div> | </div> | ||
| − | + | <div class="jcarousel-item"> | |
| − | {{banner|direction= | + | {{banner |
| + | |direction=right | ||
| + | |title = News | ||
| + | |section = [http://chuanglab.org/News PDXNet U24 grant] | ||
| + | |image=jeff-chuang-9922mirrored.jpg | ||
| + | |width=45% | ||
| + | |quote= The lab has been awarded a National Cancer Institute grant to continue our coordination of data sharing and analysis to general cancer clinical trials for the PDXNet. We are working with institutes including MD Anderson, Dana Farber, Huntsman Cancer Institute, Baylor College of Medicine, the Wistar Institute, Washington University, UC Davis, and Virginia Commonwealth University. For this project we will be analyzing hundreds of new and existing PDX samples toward the goal of developing a clinical trial based on patient-derived xenografts.}} | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 16: | Line 30: | ||
<span class="jcarousel-control-next">[[#|›]]</span> | <span class="jcarousel-control-next">[[#|›]]</span> | ||
<p class="jcarousel-pagination"></p> | <p class="jcarousel-pagination"></p> | ||
| − | </div> | + | </div> </div> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<div class="row-fluid clearfix" style="width:90%;margin:auto;text-align:center"> | <div class="row-fluid clearfix" style="width:90%;margin:auto;text-align:center"> | ||
<div class="col-md-4"> | <div class="col-md-4"> | ||
| − | <h2><span class="mw-headline" id="Research | + | <h2><span class="mw-headline" id="Research"><span class="fa fa-news"></span></span> Research</h2> |
<p>Topics that the Chuang Lab investigates | <p>Topics that the Chuang Lab investigates | ||
</p> | </p> | ||
| Line 108: | Line 47: | ||
<div class="col-md-4"> | <div class="col-md-4"> | ||
<h2><span class="mw-headline" id="Openings"><span class="fa fa"></span></span> Openings</h2> | <h2><span class="mw-headline" id="Openings"><span class="fa fa"></span></span> Openings</h2> | ||
| − | <p> | + | <p>Student/postdoctoral/research scientist openings in cancer computational biology. |
</p> | </p> | ||
<btn>Openings</btn> | <btn>Openings</btn> | ||
| Line 116: | Line 55: | ||
</p><p><br /> | </p><p><br /> | ||
</p> | </p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 11:12, 30 August 2024
Our lab uses computational and mathematical approaches to investigate the mechanisms that govern cancer cells and their spatial ecosystems. The lab specializes in problems in cancer image analysis, genomics, and evolution.
The lab uses computational and mathematical approaches to understand cancer cells and their spatial ecosystems. We develop and apply techniques from a variety of disciplines, including deep neural networks, genomic and evolutionary analysis, and biophysical modeling. We are currently focused in: 1) Multiplex and histopathological cancer image analysis using deep learning, and 2) Cancer evolution in response to therapy.
PDXNet U24 grant
The lab has been awarded a National Cancer Institute grant to continue our coordination of data sharing and analysis to general cancer clinical trials for the PDXNet. We are working with institutes including MD Anderson, Dana Farber, Huntsman Cancer Institute, Baylor College of Medicine, the Wistar Institute, Washington University, UC Davis, and Virginia Commonwealth University. For this project we will be analyzing hundreds of new and existing PDX samples toward the goal of developing a clinical trial based on patient-derived xenografts.