Difference between revisions of "Chuang Lab"
| Line 14: | Line 14: | ||
</div> | </div> | ||
<div class="jcarousel-item"> | <div class="jcarousel-item"> | ||
| − | {{banner|direction=right|title=People|section= |image= | + | {{banner|direction=right|title=People|section= |image=Gouveia_2019.jpeg|width=15%|quote=Meet the members of the Chuang lab}} |
</div> | </div> | ||
<div class="jcarousel-item"> | <div class="jcarousel-item"> | ||
Revision as of 08:51, 16 August 2019
Our lab uses computational and mathematical approaches to investigate the mechanisms that govern mammalian genomes and cell populations. The lab specializes in problems in cancer genomics, molecular evolution, and gene regulation.
The lab uses computational and mathematical approaches to understand how genomes function and evolve. We develop and apply techniques from a variety of disciplines, including molecular evolution, artificial intelligence, and biophysical modeling. We are currently focused in: 1) Cancer evolution in response to therapy, and 2) RNA-Level Gene Regulation.
Chuang Lab Awarded U24 for Data Coordination Center for NCI PDXNet
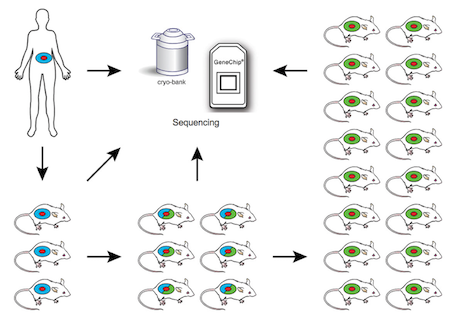
Together with Seven Bridges Genomics, we have been awarded a National Cancer Institute grant to coordinate data sharing and analysis for the PDXNet. We are working with esteemed collaborators at institutes including MD Anderson, the Huntsman Institute at the University of Utah, Baylor College of Medicine, the Wistar Institute, and Washington University. For this project we will be analyzing hundreds of new and existing PDX samples toward the goal of developing a clinical trial based on patient-derived xenografts.
Openings
As of June 2018, we have two postdoctoral openings in cancer computational biology.