Difference between revisions of "Research Topics"
| Line 3: | Line 3: | ||
Advances in sequencing have radically transformed the scale and nature of genetic studies. These have made it possible to analyze genomic changes across species, individuals, and single cells as mutations accrue and are subject to selection. Diverse phenotypic datasets have also grown rapidly, not only for sequencing-based assays such as gene expression and protein-nucleic acid interactions, but also other types including clinical and drug-screening investigations. My lab develops computational and mathematical approaches to understand how genomes function and evolve in order to make these findings clinically relevant. We use techniques from a variety of disciplines, including data science, evolutionary modeling, and biophysics. We are currently focused on two major areas: 1) Computational Approaches for Cancer Genomics, and 2) Gene Regulation. Our projects involve collaborations with experimental and computational colleagues at JAX Genomic Medicine, JAX Mammalian Genetics, and multiple outside groups. | Advances in sequencing have radically transformed the scale and nature of genetic studies. These have made it possible to analyze genomic changes across species, individuals, and single cells as mutations accrue and are subject to selection. Diverse phenotypic datasets have also grown rapidly, not only for sequencing-based assays such as gene expression and protein-nucleic acid interactions, but also other types including clinical and drug-screening investigations. My lab develops computational and mathematical approaches to understand how genomes function and evolve in order to make these findings clinically relevant. We use techniques from a variety of disciplines, including data science, evolutionary modeling, and biophysics. We are currently focused on two major areas: 1) Computational Approaches for Cancer Genomics, and 2) Gene Regulation. Our projects involve collaborations with experimental and computational colleagues at JAX Genomic Medicine, JAX Mammalian Genetics, and multiple outside groups. | ||
| + | '''Computational Approaches for Cancer Genomics'''<div> | ||
| − | + | Our lab focuses on understanding cancer using patient-derived xenografts, a model system in which human tumors are engrafted and studied in NSG mice. JAX has developed >400 such models from cancer types including breast, lung, bladder, and others, and these are a [http://tumor.informatics.jax.org/mtbwi/pdxSearch.do community wide resource]. Our lab is involved studies using these models to understand the genetic drivers of cancer and drug resistance, with a focus on tumor heterogeneity and evolution. Within JAX we work closely with other groups studying xenografts, including the Bult (JAX-MG), Liu (JAX-GM), and Lee (JAX-GM) labs. These projects include studies to identify drivers of drug susceptibility in triple negative breast cancers (Menghi et al 2016) and to determine intratumoral evolution in response to chemotherapy.<br> | |
| − | |||
| − | [[File: | + | [[File: Image:Pdx.png | 400 px]] |
| + | As of September 2017, our lab leads the NCI [https://www.pdxnetwork.org/ PDXNet] Data Commons and Coordination Center together with our colleagues at Seven Bridges Genomics. In this project, we are coordinating the analysis of novel xenograft studies across multiple centers around the United States in order to advance the development of clinical trials. Through this and other projects (Bais et al 2017), our lab has been one of the leaders in cloud computing approaches for cancer genomics analysis. <div> | ||
| + | The lab also studies evolutionary and ecological processes in a variety of other cancer systems. Recent projects have included studies of selective pressures in intratumoral evolution across thousands of cancer samples (Noorbakhsh et al 2017) and investigations into immune and stromal introgression across cancer types (Chae et al 2018). | ||
'''RNA-level Gene Regulation'''<div> | '''RNA-level Gene Regulation'''<div> | ||
Revision as of 07:05, 17 May 2018
Advances in sequencing have radically transformed the scale and nature of genetic studies. These have made it possible to analyze genomic changes across species, individuals, and single cells as mutations accrue and are subject to selection. Diverse phenotypic datasets have also grown rapidly, not only for sequencing-based assays such as gene expression and protein-nucleic acid interactions, but also other types including clinical and drug-screening investigations. My lab develops computational and mathematical approaches to understand how genomes function and evolve in order to make these findings clinically relevant. We use techniques from a variety of disciplines, including data science, evolutionary modeling, and biophysics. We are currently focused on two major areas: 1) Computational Approaches for Cancer Genomics, and 2) Gene Regulation. Our projects involve collaborations with experimental and computational colleagues at JAX Genomic Medicine, JAX Mammalian Genetics, and multiple outside groups.
Computational Approaches for Cancer GenomicsOur lab focuses on understanding cancer using patient-derived xenografts, a model system in which human tumors are engrafted and studied in NSG mice. JAX has developed >400 such models from cancer types including breast, lung, bladder, and others, and these are a community wide resource. Our lab is involved studies using these models to understand the genetic drivers of cancer and drug resistance, with a focus on tumor heterogeneity and evolution. Within JAX we work closely with other groups studying xenografts, including the Bult (JAX-MG), Liu (JAX-GM), and Lee (JAX-GM) labs. These projects include studies to identify drivers of drug susceptibility in triple negative breast cancers (Menghi et al 2016) and to determine intratumoral evolution in response to chemotherapy.
The lab also studies evolutionary and ecological processes in a variety of other cancer systems. Recent projects have included studies of selective pressures in intratumoral evolution across thousands of cancer samples (Noorbakhsh et al 2017) and investigations into immune and stromal introgression across cancer types (Chae et al 2018).
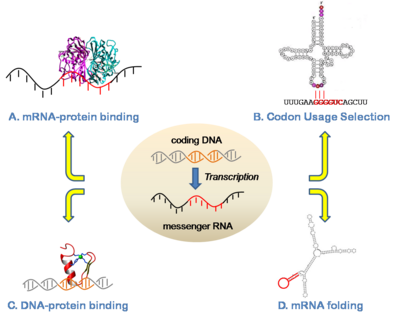
RNA-level Gene RegulationOur group has been studying mechanisms of post-transcriptional gene regulation, with focuses on regulation of translation, protein-RNA binding, and splicing. For example, in collaboration with Prof. Susan Ackerman (JAX-MG) we have identified and characterized a mutation in a tRNA as a driver for neurodegeneration and shown that this phenotype is mediated by specific translational pausing at the codons complementary to the tRNA anticodon (Ishimura et al 2014). To our knowledge, this is the first tRNA mutation found to have a phenotypic consequence in a mammal. Another current interest is how proteins interact with RNAs to achieve specific binding. In this area, we have previously developed approaches to clarify functional elements in RNA based on a combination of functional genomic, structural, and modeling approaches (Zarringhalam et al 2012). More broadly, we have been studying the functions and neutral evolutionary behavior of synonymous sites in coding sequences for more than a decade (Chuang and Li 2004; Chin et al 2005). We have shown for example that coding sequences are replete with binding sites for microRNAs, as well as other types of functional sequences such as exonic splicing enhancers. Such sites exhibit a strong selective pressure on the synonymous sites of coding regions (Kural et al 2009; Ding et al 2012; Ritter et al 2012).
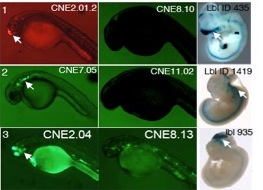
Other InterestsOne of our major prior interests has been to characterize the functions of highly conserved noncoding sequences. Just as morphological features shared among species (e.g. all vertebrates have a spine) are likely to be important to those species, DNA sequences shared among species are likely to be functional. Our lab has collaborated with the Guo lab at UCSF to study conserved noncoding elements (CNEs) via a variety of methods involving computational analysis of sequence, expression, and epigenomic data, as well as experiments testing the enhancer activity of sequences in zebrafish embryos. CNEs are abundant in vertebrate genomes, e.g. at a threshold of at least 50 bp and at least 50% sequence identity, there are 73187 strand-specific CNEs conserved between zebrafish and human. We have characterized the relative importance of cis- and trans- regulatory evolution on the functional behavior of enhancers (Ritter et al 2010) and also developed tools to organize the functions of CNEs (cneviewer.zebrafishcne.org, Persampieri et al 2008).
Our lab has been interested in a variety of issues in molecular evolution related to the balance of functional and neutral pressures in genomes. For example, one puzzle is why mutation rates are uniform in some species, such as the sensu stricto yeasts, while rates vary by location in other species, such as mouse and human. We have found that all mammalian species have regional mutation biases, typically on a scale of several megabases. In contrast, all yeasts have uniform mutation rates, with the exception of the Candida clade (Fox et al 2008; Chuang and Li 2004; Chuang and Li 2007; Chin, Chuang, and Li 2005). In species where the mutation rate is non-uniform, we are interested in questions such as what structural or sequence features affect mutation rates, and whether gene locations have evolved to make use of mutational heterogeneity.
Other model organisms with which we have expertise are the malaria parasite Plasmodium falciparum and the yeast S. cerevisiae. A central mystery of the malaria genome is how transcription is regulated. We have observed that there is far less intergenic sequence apparently under purifying selection in malaria than in yeast genomes, suggesting that transcription regulation is simpler in malaria (Imamura, Persampieri and Chuang, 2007). We have also applied comparative techniques to identify functional sites in the promoters of the Saccharomyces genus of yeasts, to estimate the complexity of gene regulation and the types of genes likely to be under the strictest regulation (Chin, Chuang, and Li 2005).
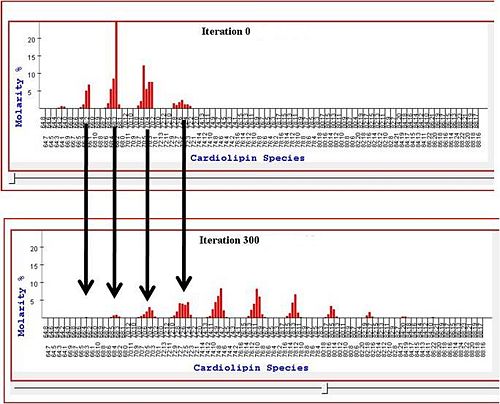
Previously the lab also has worked on the analysis of high-throughput lipidomic data. We developed tools to analyze which aspects of lipid content are important to cancer phenotypes (Kiebish et al 2008). This work is closely tied to evaluating the Warburg theory of cancer, as described in this report. We have also developed both equilibrum and dynamic models to explain the distributions of lipids found in normal and cancerous tissues (Kiebish et al 2010).