Difference between revisions of "Research Topics"
| Line 17: | Line 17: | ||
'''Gene Regulation'''<div> | '''Gene Regulation'''<div> | ||
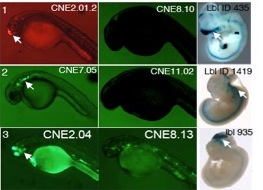
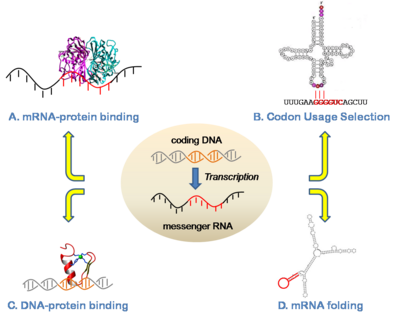
| − | Our lab studies gene regulation at both the RNA and DNA levels. For RNA, our projects have included regulation of translation, protein-RNA binding, and splicing. For example, in collaboration with Prof. Susan Ackerman (UCSD) we have identified and characterized a mutation in a tRNA as a driver for neurodegeneration and shown that this phenotype is mediated by specific translational pausing at the codons complementary to the tRNA anticodon (Ishimura et al 2014; Ishimura et al 2016). This was the first tRNA mutation found to have a phenotypic consequence in a mammal. Another current interest is how proteins interact with RNAs to achieve specific binding. In this area, we have developed approaches to identify functional elements in RNA based on functional genomic, structural, algorithmic, and high-throughput sequencing approaches (Dotu et al 2018; Zarringhalam et al 2012). Our lab has been studying the functions and neutral evolutionary behavior of synonymous sites in coding sequences for more than a decade (Chuang and Li 2004; Chin et al 2005). We have shown for example that coding sequences are replete with binding sites for microRNAs, as well as other types of functional sequences such as exonic splicing enhancers. Such sites exhibit a strong selective pressure on the synonymous sites of coding regions (Kural et al 2009; Ding et al 2012; Ritter et al 2012) | + | Our lab studies gene regulation at both the RNA and DNA levels. For RNA, our projects have included regulation of translation, protein-RNA binding, and splicing. For example, in collaboration with Prof. Susan Ackerman (UCSD) we have identified and characterized a mutation in a tRNA as a driver for neurodegeneration and shown that this phenotype is mediated by specific translational pausing at the codons complementary to the tRNA anticodon (Ishimura et al 2014; Ishimura et al 2016). This was the first tRNA mutation found to have a phenotypic consequence in a mammal. Another current interest is how proteins interact with RNAs to achieve specific binding. In this area, we have developed approaches to identify functional elements in RNA based on functional genomic, structural, algorithmic, and high-throughput sequencing approaches (Dotu et al 2018; Zarringhalam et al 2012). Our lab has been studying the functions and neutral evolutionary behavior of synonymous sites in coding sequences for more than a decade (Chuang and Li 2004; Chin et al 2005). We have shown for example that coding sequences are replete with binding sites for microRNAs, as well as other types of functional sequences such as exonic splicing enhancers. Such sites exhibit a strong selective pressure on the synonymous sites of coding regions (Kural et al 2009; Ding et al 2012; Ritter et al 2012). |
[[File:400px-Types of regulation.png]] | [[File:400px-Types of regulation.png]] | ||
Revision as of 18:03, 8 April 2021
Broad advances in sequencing and imaging have radically transformed the nature of computational biology. Many datatypes are rapidly increasing -- including imaging, RNA, DNA, protein-DNA binding, clinical, and drug-screening data -- providing rich avenues for discovery at the interface of mechanistic modeling and machine learning. My lab uses computational and mathematical approaches to understand how cell populations function, how they evolve, and to predict their behavior under drug treatment. Our work combines disciplines including machine learning, sequence analysis, molecular evolution and biophysical modeling. We are currently focused in three major areas: 1) Cancer Evolution and Patient-Derived Xenografts, 2) Cancer Image Analysis, and 2) Gene Regulation. These projects involve collaborations with experimental and computational colleagues at JAX Genomic Medicine, JAX Mammalian Genetics, and a number of outside groups.
For the most up-to-date information of the lab's work, see the Publications page.
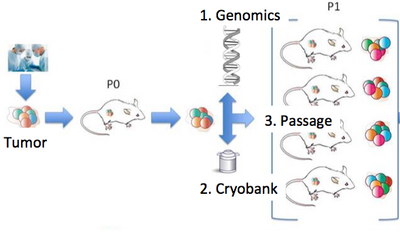
Cancer Evolution and Patient-Derived XenograftsOur lab is a leader in the field of patient-derived xenografts, a model system in which human tumors are engrafted and studied in NSG mice. JAX has developed >300 such models from cancer types including breast, lung, bladder, and others, and these are a community wide resource. Since 2017, our lab co-leads the Data Coordination Center for the NCI PDXNet, a multi-institute consortium supported by the NCI Cancer Moonshot Initative. We are partnering with institutes around the country including the Huntsman Cancer Institute, Baylor College of Medicine, MD Anderson, Washington University, the Wistar Institute, NCI's Frederick National Laboratory, and Seven Bridges Genomics to jointly study cancer using xenografts. Our lab uses these to understand the genetic drivers of cancer and drug resistance, with a focus on computational modeling of intratumoral evolution (Woo, Giordano et al 2021, Kim et al 2018, Noorbakhsh and Chuang 2017) and also the robustness of xenografts for preclinical testing therapeutic agents (Evard et al 2020). For more information see the PDXNetwork site.
Within JAX we work closely with a number of groups studying cancer with xenografts and clinical samples, including the Liu, Bult, Lee, Palucka, Robson, and Anczukow-Camarda labs, on projects such as triple negative breast cancer (Menghi et al 2016), gastric cancer (Cho et al 2018), and immune cell activity (Bais et al 2017; Chae et al 2018).
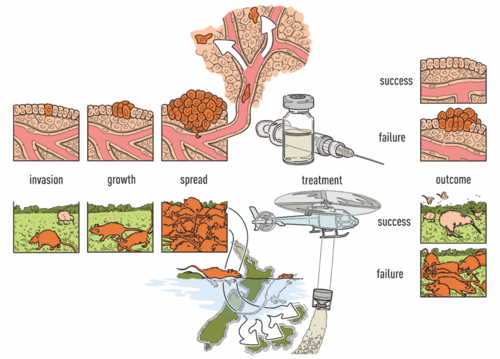
Cancer Image AnalysisCancer treatment response and resistance are affected by the distribution of tumor subclones, immune cells, and other features of the microenvironment. To understand this web of interactions, the lab is developing approaches for computational image analysis, an approach that can reveal the spatial processes driving intratumoral heterogeneity. For example, we have recently developed convolutional neural networks that can be applied to H&E images to distinguish tumor from normal tissue with 99% accuracy, identify cancer subtypes, and classify cancers having particular mutations (Noorbakhsh, Farahmand, Foroughi Pour et al 2020). These works have shown that networks trained on one cancer type have predictive power in other cancer types, indicating that combinatorial approaches can be used in image analysis for discovery science. The lab analyzes many other types of imaging data as well, including histocytometry, IHC, and imaging mass cytometry for tumor samples, as well as confocal imaging of organoids. The lab is also broadly interested in ecological modeling of cancer, which has remarkable similarities to the eradication of invasive pests from islands in the New Zealand archipelago (Noorbakhsh, Zhao, Russell, and Chuang 2019)
Gene RegulationOur lab studies gene regulation at both the RNA and DNA levels. For RNA, our projects have included regulation of translation, protein-RNA binding, and splicing. For example, in collaboration with Prof. Susan Ackerman (UCSD) we have identified and characterized a mutation in a tRNA as a driver for neurodegeneration and shown that this phenotype is mediated by specific translational pausing at the codons complementary to the tRNA anticodon (Ishimura et al 2014; Ishimura et al 2016). This was the first tRNA mutation found to have a phenotypic consequence in a mammal. Another current interest is how proteins interact with RNAs to achieve specific binding. In this area, we have developed approaches to identify functional elements in RNA based on functional genomic, structural, algorithmic, and high-throughput sequencing approaches (Dotu et al 2018; Zarringhalam et al 2012). Our lab has been studying the functions and neutral evolutionary behavior of synonymous sites in coding sequences for more than a decade (Chuang and Li 2004; Chin et al 2005). We have shown for example that coding sequences are replete with binding sites for microRNAs, as well as other types of functional sequences such as exonic splicing enhancers. Such sites exhibit a strong selective pressure on the synonymous sites of coding regions (Kural et al 2009; Ding et al 2012; Ritter et al 2012).
Much of the lab's research has grown out of early interests in molecular evolution and statistical physics. In some of our earlier work, we have characterized the relative importance of cis- and trans- regulatory evolution on enhancers (Ritter et al 2010; Persampieri et al 2008). We have also studied the evolution of mutational processes across species and cancers. This has included research into why mutation rates are uniform in some species, such as the sensu stricto yeasts, while rates vary by location in other species, such as mouse and human (Fox et al 2008; Chuang and Li 2004; Chuang and Li 2007; Chin, Chuang, and Li 2005). Biophysics interests have included the dynamics of translocation of a polymer through a nanopore (Chuang et al, Phys Rev E 2001) and the thermodynamic stability of protein folds (Chuang et al, Phys Rev Lett 2001).